Sparse Networks with Overlapping Communities (SNetOC) package: demo_sparsity
This Matlab script shows empirically the sparsity properties of a range of graph models.
For downloading the package and information on installation, visit the SNetOCC webpage.
Reference:
- A. Todeschini, X. Miscouridou and F. Caron (2017) Exchangeable Random Measures for Sparse and Modular Graphs with Overlapping Communities. arXiv:1602.02114.
Authors:
- A. Todeschini, Inria
- X. Miscouridou, University of Oxford
- F. Caron, University of Oxford
Tested on Matlab R2016a. Requires the Statistics toolbox.
Last Modified: 10/10/2017
Contents
General settings
clear close all % Add paths addpath ./GGP/ ./CGGP/ ./utils/ % Save plots and workspace saveplots = false; saveworkspace = true; root = '.'; outpath = fullfile(root, 'results', 'demo_sparsity', date); if (saveplots || saveworkspace) && ~isdir(outpath) mkdir(outpath) end % Set the seed rng default
Definition of the graph models
p = 2; alpha = 100; tau = 1; gamma = zeros(p,1); a = .2; b = 1/p; Fdist.name = 'gamma'; Fdist.param.a = a; Fdist.param.b = b; typegraph = 'undirected'; observe_all = false; ntrials = 500; nsamples = 1; imod = 0; % CGGP (sigma=-0.5) imod = imod+1; sigma = -0.5; obj{imod} = graphmodel('CGGP', p, alpha, sigma, tau, Fdist, gamma, typegraph, observe_all); field{imod} = 'alpha'; trial{imod} = linspace(1, 1500, ntrials); optionrnd{imod} = 1e-10; % CGGP (sigma=0.2) imod = imod+1; sigma = 0.2; obj{imod} = graphmodel('CGGP', p, alpha, sigma, tau, Fdist, gamma, typegraph, observe_all); field{imod} = 'alpha'; trial{imod} = linspace(1, 600, ntrials); optionrnd{imod} = 1e-10; % CGGP (sigma=0.5) imod = imod+1; sigma = 0.5; obj{imod} = graphmodel('CGGP', p, alpha, sigma, tau, Fdist, gamma, typegraph, observe_all); field{imod} = 'alpha'; trial{imod} = linspace(1, 300, ntrials); optionrnd{imod} = 1e-5; % CGGP (sigma=0.8) imod = imod+1; sigma = 0.8; obj{imod} = graphmodel('CGGP', p, alpha, sigma, tau, Fdist, gamma, typegraph, observe_all); field{imod} = 'alpha'; trial{imod} = linspace(1, 300, ntrials); optionrnd{imod} = 1e-4; edgebins = 2.^(0:1:12); sizebins = edgebins(2:end) - edgebins(1:end-1); sizebins(end+1) = 1;
Sample graphs of various sizes
for k=1:numel(obj) % For different models fprintf('--- Model %d/%d: %s ---\n', k, numel(obj), obj{k}.name) for i=1:numel(trial{k}) % Varying number of nodes if rem(i, 10)==0 fprintf('Trial %d/%d \n', i, numel(trial{k})); end obj{k}.param.(field{k}) = trial{k}(i); for j=1:nsamples % For different samples G = graphrnd(obj{k}, optionrnd{k}); % Sample the graph degreefreq{k}(i,j,:) = zeros(numel(edgebins), 1); if size(G, 1)>0 degreefreq{k}(i,j,:) = histc(full(sum(G)),edgebins)./sizebins/size(G, 1); end nbnodes{k}(i,j) = size(G, 1); nbedges{k}(i,j) = sum(G(:))/2 + trace(G)/2; maxdegree{k}(i,j) = max(sum(G)); degreeone{k}(i,j) = sum(sum(G)==1); end end end
--- Model 1/4: Compound generalized gamma process --- Trial 10/500 Trial 20/500 Trial 30/500 Trial 40/500 Trial 50/500 Trial 60/500 Trial 70/500 Trial 80/500 Trial 90/500 Trial 100/500 Trial 110/500 Trial 120/500 Trial 130/500 Trial 140/500 Trial 150/500 Trial 160/500 Trial 170/500 Trial 180/500 Trial 190/500 Trial 200/500 Trial 210/500 Trial 220/500 Trial 230/500 Trial 240/500 Trial 250/500 Trial 260/500 Trial 270/500 Trial 280/500 Trial 290/500 Trial 300/500 Trial 310/500 Trial 320/500 Trial 330/500 Trial 340/500 Trial 350/500 Trial 360/500 Trial 370/500 Trial 380/500 Trial 390/500 Trial 400/500 Trial 410/500 Trial 420/500 Trial 430/500 Trial 440/500 Trial 450/500 Trial 460/500 Trial 470/500 Trial 480/500 Trial 490/500 Trial 500/500 --- Model 2/4: Compound generalized gamma process --- Trial 10/500 Trial 20/500 Trial 30/500 Trial 40/500 Trial 50/500 Trial 60/500 Trial 70/500 Trial 80/500 Trial 90/500 Trial 100/500 Trial 110/500 Trial 120/500 Trial 130/500 Trial 140/500 Trial 150/500 Trial 160/500 Trial 170/500 Trial 180/500 Trial 190/500 Trial 200/500 Trial 210/500 Trial 220/500 Trial 230/500 Trial 240/500 Trial 250/500 Trial 260/500 Trial 270/500 Trial 280/500 Trial 290/500 Trial 300/500 Trial 310/500 Trial 320/500 Trial 330/500 Trial 340/500 Trial 350/500 Trial 360/500 Trial 370/500 Trial 380/500 Trial 390/500 Trial 400/500 Trial 410/500 Trial 420/500 Trial 430/500 Trial 440/500 Trial 450/500 Trial 460/500 Trial 470/500 Trial 480/500 Trial 490/500 Trial 500/500 --- Model 3/4: Compound generalized gamma process --- Trial 10/500 Trial 20/500 Trial 30/500 Trial 40/500 Trial 50/500 Trial 60/500 Trial 70/500 Trial 80/500 Trial 90/500 Trial 100/500 Trial 110/500 Trial 120/500 Trial 130/500 Trial 140/500 Trial 150/500 Trial 160/500 Trial 170/500 Trial 180/500 Trial 190/500 Trial 200/500 Trial 210/500 Trial 220/500 Trial 230/500 Trial 240/500 Trial 250/500 Trial 260/500 Trial 270/500 Trial 280/500 Trial 290/500 Trial 300/500 Trial 310/500 Trial 320/500 Trial 330/500 Trial 340/500 Trial 350/500 Trial 360/500 Trial 370/500 Trial 380/500 Trial 390/500 Trial 400/500 Trial 410/500 Trial 420/500 Trial 430/500 Trial 440/500 Trial 450/500 Trial 460/500 Trial 470/500 Trial 480/500 Trial 490/500 Trial 500/500 --- Model 4/4: Compound generalized gamma process --- Trial 10/500 Trial 20/500 Trial 30/500 Trial 40/500 Trial 50/500 Trial 60/500 Trial 70/500 Trial 80/500 Trial 90/500 Trial 100/500 Trial 110/500 Trial 120/500 Trial 130/500 Trial 140/500 Trial 150/500 Trial 160/500 Trial 170/500 Trial 180/500 Trial 190/500 Trial 200/500 Trial 210/500 Trial 220/500 Trial 230/500 Trial 240/500 Trial 250/500 Trial 260/500 Trial 270/500 Trial 280/500 Trial 290/500 Trial 300/500 Trial 310/500 Trial 320/500 Trial 330/500 Trial 340/500 Trial 350/500 Trial 360/500 Trial 370/500 Trial 380/500 Trial 390/500 Trial 400/500 Trial 410/500 Trial 420/500 Trial 430/500 Trial 440/500 Trial 450/500 Trial 460/500 Trial 470/500 Trial 480/500 Trial 490/500 Trial 500/500
Some plots
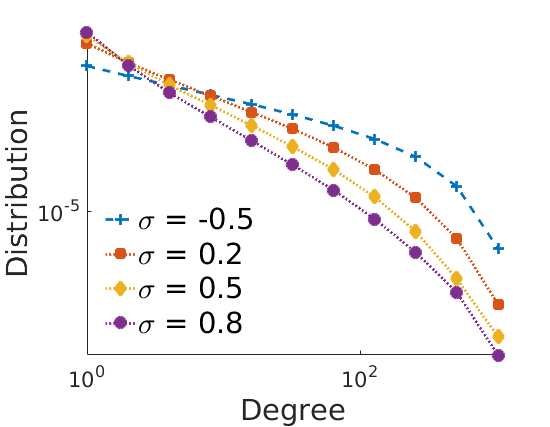
% Properties of the plots fontsize = 22; plotstyles = {'--+',':s',':d',':o'}; colors = lines; for k=1:numel(obj) leg{k} = sprintf('\\sigma = %g', obj{k}.param.sigma); end set(0,'DefaultAxesFontSize', 16) set(0,'DefaultTextFontSize', fontsize) set(0,'DefaultLineLineWidth', 2) set(0,'DefaultLineMarkerSize', 8) close % Degree distribution loglog plot figure('name', 'degreeloglog') for k=1:numel(obj) h = loglog(edgebins, squeeze(mean(mean(degreefreq{k}, 1), 2)), plotstyles{k}); set(h, 'markerfacecolor', colors(k,:), 'color', colors(k,:)); hold on end axis tight % xlim([1, 300]) % ylim([10, 20000]) xlabel('Degree', 'fontsize', fontsize) ylabel('Distribution', 'fontsize', fontsize) legend(leg, 'fontsize', fontsize, 'location', 'southwest') legend boxoff box off if saveplots savefigs(gcf, 'degree', outpath); end % Nb of Edges vs nb of nodes on loglog plot step = .4; figure('name', 'edgesvsnodesloglog'); for k=1:numel(obj) h = plot_loglog(nbnodes{k}(:), nbedges{k}(:), plotstyles{k}, step); set(h, 'markerfacecolor', colors(k,:), 'color', colors(k,:)); hold on end axis tight xlim([25, 3000]) % ylim([10, 20000]) xlabel('Number of nodes', 'fontsize', fontsize) ylabel('Number of edges', 'fontsize', fontsize) legend(leg, 'fontsize', fontsize, 'location', 'northwest') legend boxoff box off if saveplots savefigs(gcf, 'edgesvsnodes', outpath); end
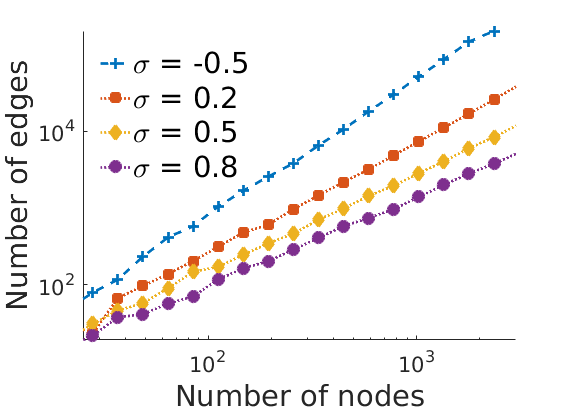
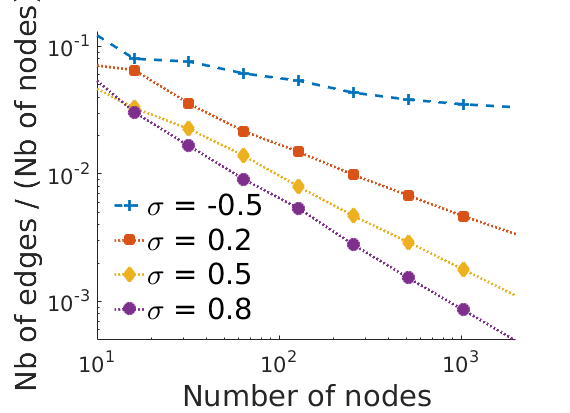
% Nb of Edges/Nb of nodes squared vs nb of nodes on loglog plot step = 1; figure('name', 'edgesvsnodesloglog') for k=1:numel(obj) ind = nbnodes{k}(:)>0; h = plot_loglog(nbnodes{k}(ind), nbedges{k}(ind)./nbnodes{k}(ind).^2, plotstyles{k}, step); set(h, 'markerfacecolor', colors(k,:), 'color', colors(k,:)); hold on end axis tight xlim([10, 2000]) xlabel('Number of nodes', 'fontsize', fontsize) ylabel('Nb of edges / (Nb of nodes)^2', 'fontsize', fontsize) legend(leg,'fontsize', fontsize, 'location', 'southwest') legend boxoff box off if saveplots savefigs(gcf, 'edgesvsnodes2', outpath); end
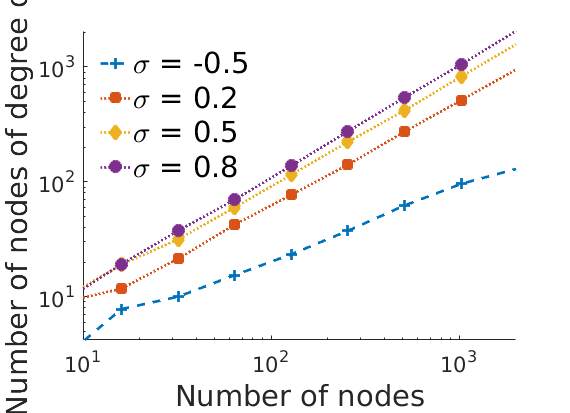
% Nb of nodes of degree one versus number of nodes figure('name', 'degonevsnodes'); for k=1:numel(obj) h = plot_loglog(nbnodes{k}(:), degreeone{k}(:), plotstyles{k}, step); set(h, 'markerfacecolor', colors(k,:), 'color', colors(k,:)); hold on end axis tight xlim([10, 2000]) % ylim([1, 2000]) xlabel('Number of nodes', 'fontsize', fontsize); ylabel('Number of nodes of degree one', 'fontsize', fontsize); legend(leg,'fontsize', fontsize, 'location', 'northwest'); legend('boxoff'); legend boxoff box off if saveplots savefigs(gcf, 'degreeonevsnodes', outpath); end
Save workspace
if saveworkspace save(fullfile(outpath, 'workspace')); end